Table of Contents
The Grand Beginnings
The Human Genome Project, completed in the early 2000s, marked the beginning of an era of bioinformatics research. It was a remarkable endeavor to sequence and map the first complete human genome. Lasting more than a decade, involving thousands of scientists worldwide, and costing a whopping 2.7 billion dollars, it was a small price for the avalanche it triggered. From project’s completion until today we sequenced more than a thousand species, annotated over 250 million genes, discovered millions of mutations, and identified thousands of rare diseases.
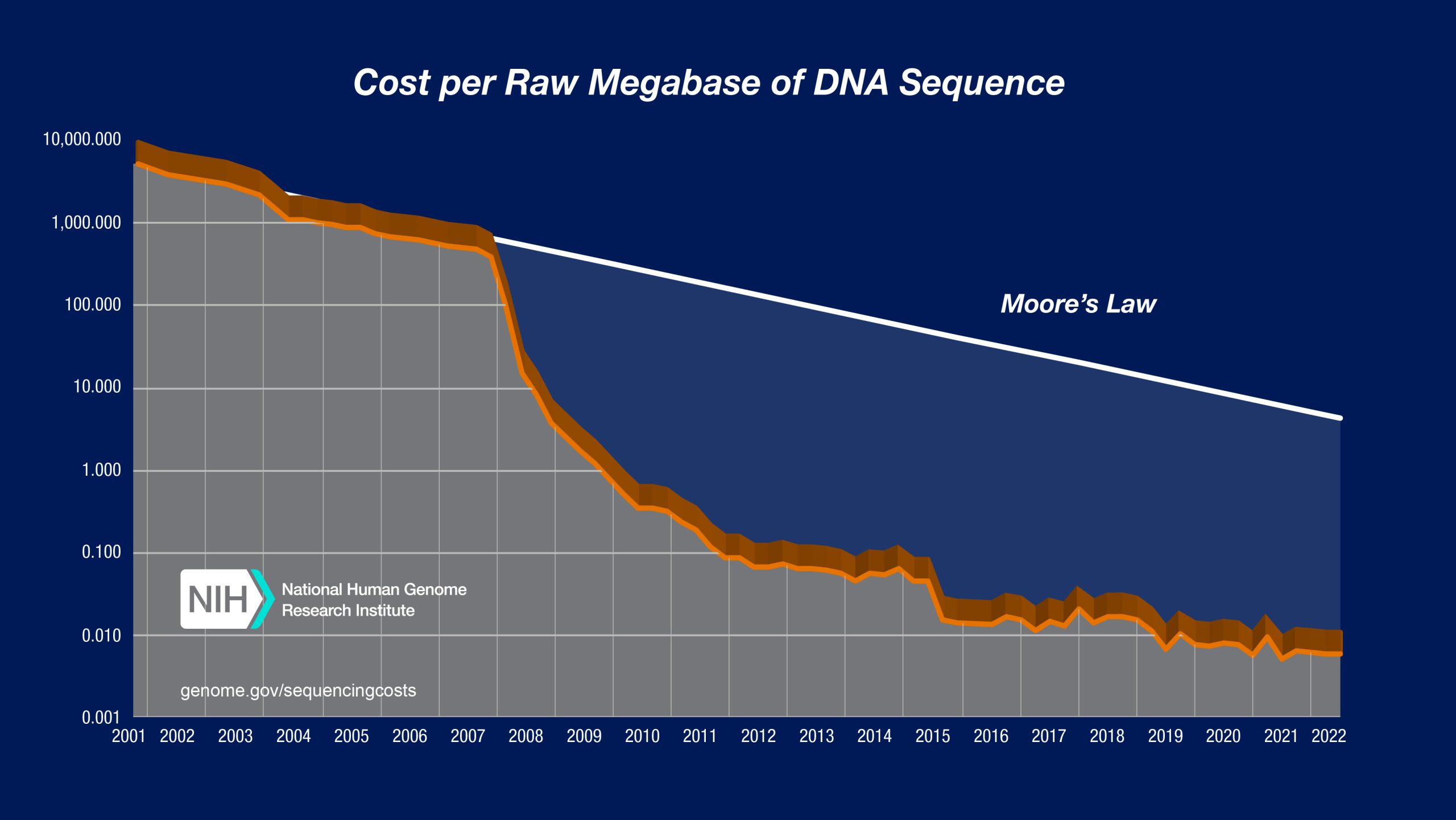
The project brought about the dawn of genomics, phylogenetics, epigenetics, proteomics, microbiome research, and personalized medicine, among others, all which led to technologies such as the fourth generation of high-throughput sequencing, together with advanced analytical algorithms and computational methods. This revolution impacted every branch of Biology and Medicine. Today, in just over 20 years, we are able to sequence a human genome in a day for the price of an airline ticket — a million-fold reduction in cost and effort (Figure 1). And yet, we understand only a fraction of it [1].
Challenges in Bioinformatics
New and affordable sequencing technologies can produce more data than we can presently analyze. In other words, despite the great advancements, our analytical methods and computational infrastructure are lagging behind. Since the onset of the COVID-19 pandemic until today, we sequenced 16.5 million SARS-CoV-2 genomes [3]. Whether this extensive sequencing effort was a panic driven overreaction, is a question for another time. However, this large influx of data presented bioinformaticians, programmers and data analysts with challenges they haven’t faced before, regardless of the viral genome being 200,000 times smaller than the human genome. The challenges ranged from sampling biases, issues with data integration, data mining, and scattered resources to problems with data transfer, and scaling of our analytical methods [4], [5].
Somewhat surprisingly, we are still not able to utilize this vast sequencing data to its full potential. For example, 20 years after the sequencing of the first human genome, we still don’t know the exact number of human genes [6]. The number revolves around some 20,000, yet, these protein-coding genes represent only about 1% of the entire genomic sequence. The exact function of the remaining genomic ‘dark-matter’, which is comprised of elements such as low-complexity regions, introns, regulatory elements, pseudogenes, microRNAs and long non-coding RNAs, remains largely unknown.
But, let’s move beyond humans and back to one of the smallest biological entities – viruses. SARS-CoV-2 is a zoonotic virus, meaning it changed its host from an animal to a human. Estimates show there are more than 1.5 million different mammalian and waterfowl viruses, of which 631,000 to 827,000 are zoonotic and can potentially infect humans [7]. At present, we have some 12,000 complete viral genomes in our databases. Let’s pretend all these genomes belong to zoonotic viruses (which they clearly do not). Even this broad generalization implies that we are unaware of more than 98% of the viruses that are a potential threat to the global health. Take a moment to ponder upon that.
The Way Forward
Over the past 20 years, bioinformatics has primarily remained an academic discipline. As such, it could not deliver standardized data practices, optimized workflows, user-friendly software, robust predictive models, or unified database resources. I picture it as a foamy structure resembling multitude of soap bubbles lacking a central surface to coalesce around. For instance, you have phylogenomic research, drug–target interaction prediction, or spatial transcriptomics, and world of knowledge and resources which sets them apart.
Perhaps a change is on the horizon. In the coming years, we may see a more cohesive, accessible, and robust bioinformatics. The increasing affordability of sequencing technologies mentioned earlier (Figure 2) is pushing bioinformatics toward a sustainable business model. There are now companies offering services rooted in bioinformatics research which range from ancestry analyses, personalized medicine, assessment of hereditary diseases, nutrition, metabolism, microbiome and more. I feel the evolution of bioinformatics mirrors that of the field of machine learning which transitioned from a multitude of statistical methods scattered across scientific journals to AI becoming a core business model of leading IT companies.
There are oceans of biological data still uncharted and this is where the Omics Solutions’ ship is sailing… metaphorically of course. We have not purchased any ships… yet. However, with or without a ship, our mission remains – to take charge of innovation in bioinformatics and to tackle the burning issues across the life sciences. Similar to AI and machine learning, we want to help bioinformatics take that leap and pave the way for groundbreaking scientific discoveries and developments.
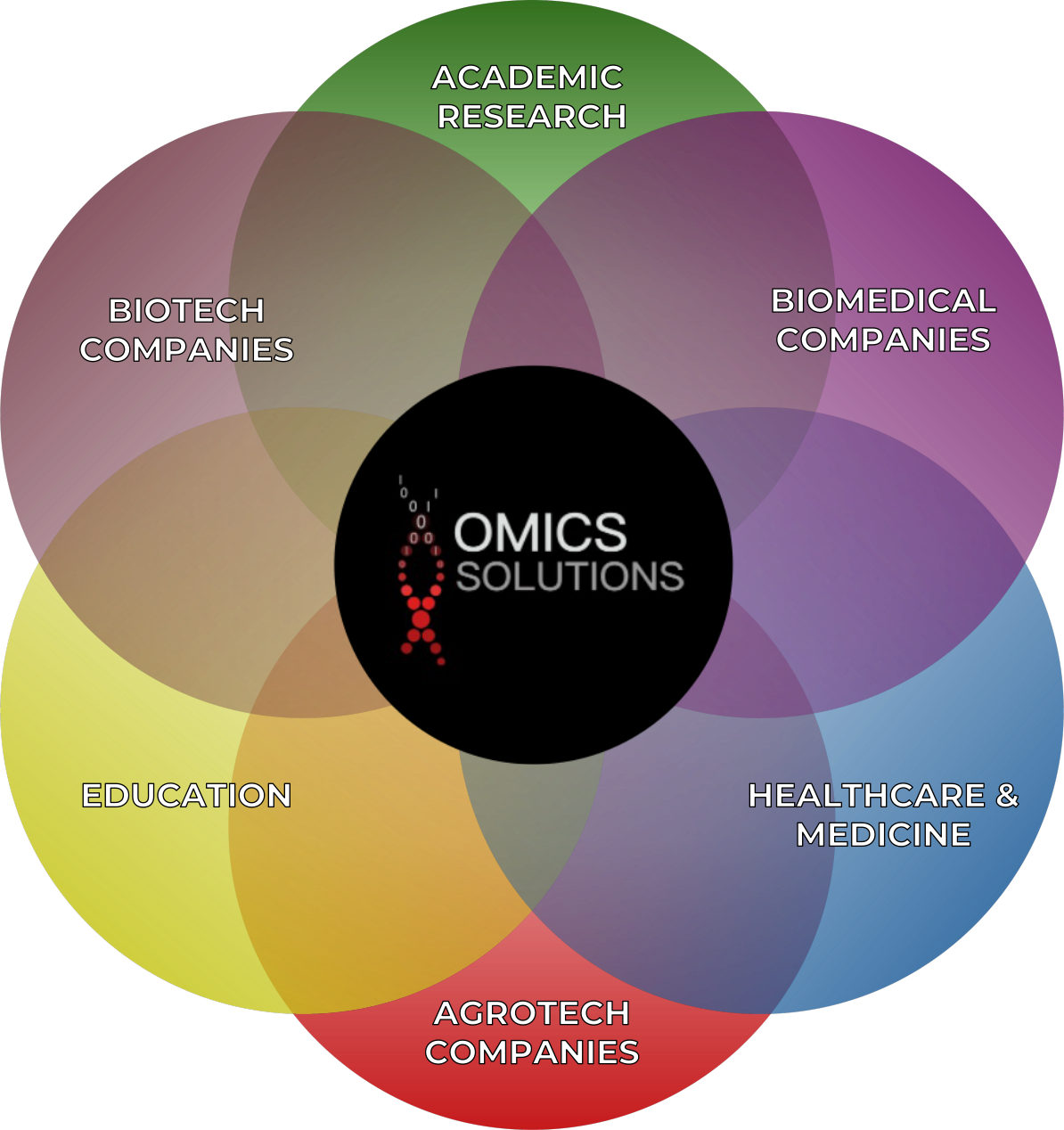
At the time of writing, our team commands the skills and knowledge needed to tackle challenges across industries and various areas of research (Figure 3). Some examples include genome wide association studies (GWAS), biomarker discoveries, de novo assemblies and genome mapping, data workflow integration, genome annotation, functional predictions, metagenome analysis, single cell, spatial or bulk transcriptomics, analysis of methylation patterns, proteomics and protein function prediction, meta-analyses, building predictive models, network analysis, omics integration, and many more. As our team grows so will our knowledge and analytical capabilities.
If you haven’t already, I invite you to have a look at our graphical interface where you can browse through our skills and expertise. If you have, maybe you can join our mailing list and we will let you know when the next cruise on our ship is?
P.S. And that image at the top… The books labeled with numbers from 1 to 23. That is a single human genome printed on paper.
References
[1] R. Guigó, “Genome annotation: From human genetics to biodiversity genomics,” Cell Genomics, vol. 3, no. 8, p. 100375, Aug. 2023, doi: 10.1016/J.XGEN.2023.100375.
[2] NHGRI, “DNA Sequencing Costs: Data.” Accessed: Feb. 29, 2024. [Online]. Available: https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data
[3] “GISAID – gisaid.org.” Accessed: Mar. 01, 2024. [Online]. Available: https://gisaid.org/
[4] S. Pal, S. Mondal, G. Das, S. Khatua, and Z. Ghosh, “Big data in biology: The hope and present-day challenges in it,” Gene Rep, vol. 21, p. 100869, Dec. 2020, doi: 10.1016/J.GENREP.2020.100869.
[5] J. K. Chaudhari, S. Pant, R. Jha, R. K. Pathak, and D. B. Singh, “Biological big-data sources, problems of storage, computational issues, and applications: a comprehensive review,” Knowledge and Information Systems 2024, pp. 1–51, Jan. 2024, doi: 10.1007/S10115-023-02049-4.
[6] P. Amaral et al., “The status of the human gene catalogue,” Nature 2023 622:7981, vol. 622, no. 7981, pp. 41–47, Oct. 2023, doi: 10.1038/s41586-023-06490-x.
[7] O. Jonas and R. Seifman, “Do we need a Global Virome Project?,” Lancet Glob Health, vol. 7, no. 10, pp. e1314–e1316, Oct. 2019, doi: 10.1016/S2214-109X(19)30335-3.
[8] “File:Oxford nanopore MinION flow cell back.jpg – Wikimedia Commons.” Accessed: Mar. 04, 2024. [Online]. Available: https://commons.wikimedia.org/wiki/File:Oxford_nanopore_MinION_flow_cell_back.jpg