Table of Contents
On the Importance of Gene Expression Analysis
Though it may appear to be a narrow field, biology is an ocean, and bioinformatics research is a great river feeding its delta into that vast expanse. One branch of this delta aims to understand the intricate layers of gene regulation and expression, which are crucial for advancing research, clinical applications, and food production alike. In its purest form, this branch is known as gene expression analysis.
In bioinformatics, especially with the advent of next-generation sequencing technologies, gene expression analysis has taken center stage. It encompasses all forms of statistical analyses performed on data generated by sequencing the expression profiles of individual cells or bulk tissue samples. This sequencing process, known as RNA sequencing (RNA-seq), is why such analysis is often referred to as RNA-seq analysis. Just as a photograph captures information about the action taking place behind the scenes, RNA molecules carry information about the “action” occurring inside a living cell. Therefore, RNA-seq analysis can be thought of as a tool for examining the activity within an organism, a tissue, or a single cell.
Gene expression analysis enables researchers to examine which genes are active, which variants of those genes are present, what signals or mutations they carry, when the genes are active, and to what extent across different tissues, cell types, or under varying conditions. RNA-seq is a powerful tool that provides a high-resolution, quantitative snapshot of gene activity, making it possible to uncover insights into development, disease states, and cellular responses, among others.
Despite its potential, RNA-seq-based analyses come with certain trade-offs and challenges. These include handling large matrices and massive data files, complex batch effects and normalization procedures, lack of benchmarking standards, incompatibility of data across projects and sequencing technologies, and multi-layered statistical models. Tackling these challenges—along with developing efficient data pipelines, intuitive visualizations, and user-friendly interfaces—is essential for making RNA-seq more accessible to researchers. One of Omics’ clients set out to do exactly that: create a gene expression analysis platform.
An Overview of the Gene Expression Analysis Platform (GEAP)
Omics was proud to be at the core of GEAP’s development, providing both scientific and technical leadership throughout the creation of the platform. From designing the analytical backbone and managing custom projects to building interactive visualization interfaces and developing novel proprietary algorithms, we helped turn a bold vision into a powerful and accessible tool for researchers and diverse stakeholders.
The idea behind GEAP was simple: to harness, scale up, and streamline the power of gene expression insights for the benefit of the food production and research efforts. More specifically, GEAP was built to optimize the productivity of farm animals through precision nutrition and health programs. The platform itself is conceived as cloud-based and is structured around two modules: the first one being tailored for researchers, while the second is designed to be more accessible to farmers, offering direct recommendations and actionable insights.
Depending on their specific needs, users can choose to either upload their own data or request a sample collection protocol. In the latter case, they will receive a sampling kit along with detailed instructions on how to collect the samples and ship them to a sequencing laboratory. Upon receiving the data and the necessary user input, the platform begins processing and analyzing the information, displaying results and reports in real time as they are generated.

The Data Analysis Pipeline Design
The preprocessing of raw RNA sequencing files follows a well-established trajectory: quality control, trimming, alignment, and quantification. The raw data, along with metadata, QC reports, and gene counts, are stored in a database accessible via the platform. Once preprocessing is successfully completed, the user is notified via email and can choose to proceed with one of the available analysis modules. RNA-seq analysis operates on gene counts using proprietary pipelines built from scratch. Key features include:
- Batch correction and normalization
- Differential gene expression analysis
- Gene set enrichment analysis
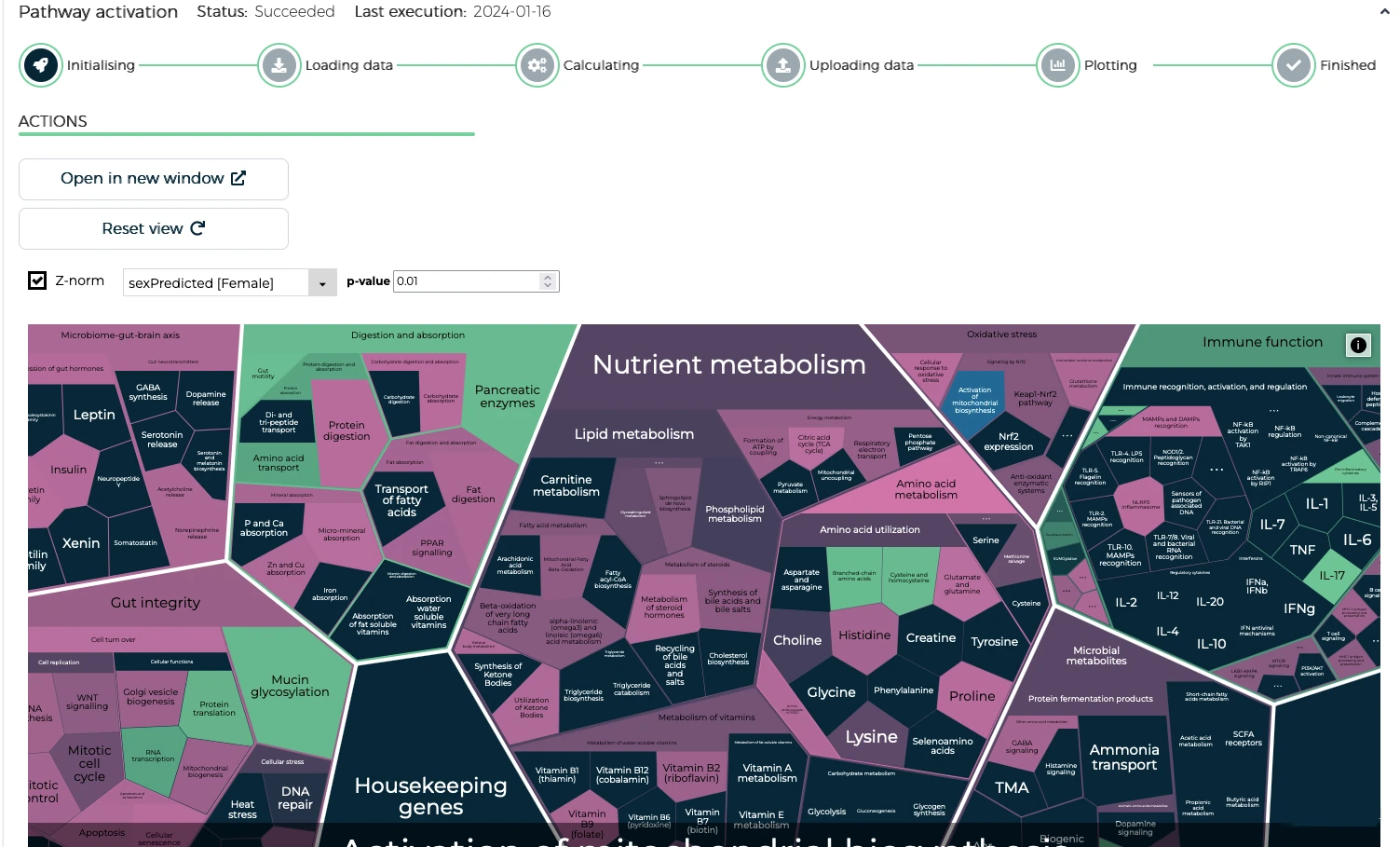
- Pathway activation analysis
- Biomarker discovery
- Machine learning (ML) predictive models
The results from each module is displayed on the platform in an interactive format. Users can seamlessly explore the data down to the finest details using a rich set of visualizations. They can generate reports and access or download their data, analysis logs, and tables.
Scientific Output and Real-World Impact
Our work on the platform exemplifies Omics’ broader mission: to empower researchers with intuitive, scalable, and reproducible tools for high-dimensional biological data. Contribution of Omics team spanned all aspects of development-from project coordination and bioinformatics design to backend infrastructure and software architecture – achieved through close collaboration with software engineers and backend developers. We successfuly built a robust and accessible RNA-seq analysis platform with some very impactful features, such as:
- Scalable cloud infrastructure designed to efficiently handle petabytes of gene expression data.
- Automated and intuitive end-to-end data analysis pipelines with minimal user interventions required.
- Interactive visualization tools specifically tailored for biological interpretation and actionable insights.
- Secure data storage and sharing capabilities.
- Proprietary and novel algorithms and analytical frameworks.
Several research articles have emerged from this initiative. These publications leveraged platform’s infrastructure for RNA-seq analysis and visualization, demonstrating the platform’s prowess through a peer-review process. Today, the platform continues to operate successfully, expanding its user base and driving new scientific discoveries.

