Table of Contents
Therapeutic Promise of ncRNAs
Non-coding RNAs (ncRNAs), especially long non-coding RNAs (lncRNAs), have moved from overlooked genome elements to important subjects in biomedical research. They are strongly implicated in a broad spectrum of diseases, including cancer, autoimmune conditions, and neurodegeneration.
Although they do not encode proteins, lncRNAs can influence chromatin state, transcription, splicing, RNA stability, translation, and the assembly of protein complexes. These regulatory activities are conditional upon cell type–specific expression of the lncRNA, correct nuclear or cytoplasmic localization, and co-localization with essential binding partners. Their functional complexity along with limited annotation make them challenging to study, which requires careful and comprehensive analysis. Unlocking their potential as biomarkers or therapeutic targets calls for a strategic, multidisciplinary approach that can synthesize diverse sources of evidence into meaningful conclusions.
Types and Functions of Regulatory ncRNAs
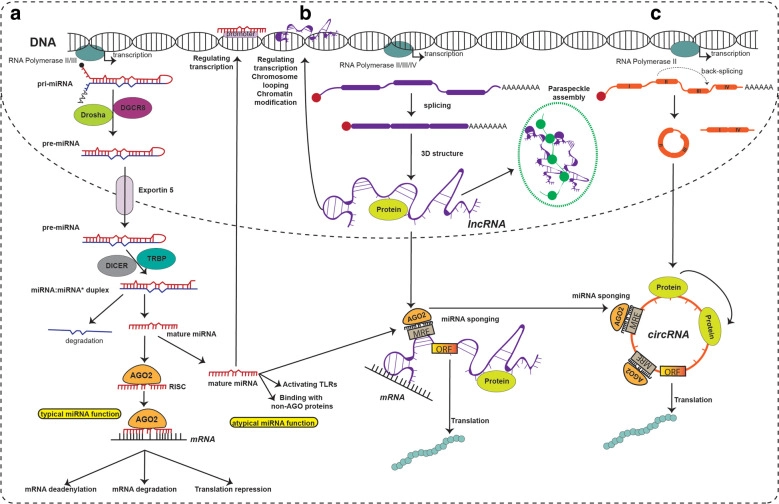
ncRNAs are RNA molecules that are not translated into proteins but play critical roles in gene regulation and cellular function. They are broadly classified into housekeeping and regulatory ncRNAs, of which the latter have a therapeutic potency.
Namely, these are:
- miRNAs (microRNAs): ~22 nt; post-transcriptionally repress gene expression by binding target mRNAs.
- lncRNAs (long non-coding RNAs): >200 nt; regulate transcription, chromatin structure, mRNA stability, protein function, and act as scaffolds or decoys.
- circRNAs (circular RNAs): Covalently closed loops formed by back-splicing; act as miRNA sponges, protein decoys, scaffolds, or encode micropeptides.
- piRNAs (PIWI-interacting RNAs): Regulate transposable elements, mostly in germ cells.
- siRNAs (small interfering RNAs): Involved in RNA interference; silence specific genes.
- eRNAs (enhancer RNAs): Transcribed from enhancer regions; promote gene transcription.
Among these, miRNAs, lncRNAs, and circRNAs are particularly important due to their diverse regulatory roles in development, cell differentiation, immune response, and disease. They also form regulatory networks, often interacting with each other (e.g., lncRNAs and circRNAs sponging miRNAs), adding layers of complexity to gene regulation.
Picture taken from: Chen, B., Dragomir, M.P., Yang, C. et al. Targeting non-coding RNAs to overcome cancer therapy resistance. Sig Transduct Target Ther 7, 121 (2022). https://doi.org/10.1038/s41392-022-00975-3
Framework for ncRNA Target Discovery
As mentioned, non-coding RNAs include a wide range of RNA types which differ significantly in size, function, and biogenesis pathways. This diversity means that a single workflow cannot address all ncRNA types effectively. Therefore, we adopt a modular strategy tailored to each class and biological context. By assessing an ncRNA’s unique interaction network, we select the most appropriate data types and analytical methods.
This begins with a deep dive into the existing literature: mapping what is known, identifying recent advancements in the field, cataloguing available resources, and pinpointing the gaps. This enables us to build an informed analytical strategy. The next step involves identifying both established and emerging analytical approaches which will help us understand how different layers of data might be leveraged in the target discovery.
Furthermore, available databases and repositories are explored to support the findings, provide context, and reinforce their biological relevance. By integrating findings from existing studies, data-driven analysis, and curated biological knowledge, a more confident and meaningful shortlist of candidate targets can be formed.
This integrative approach ensures that every discovery is founded on both data and existing experimental evidence, while also allowing for automation and scalability.
Custom-Built Discovery Platform
To meet the complexity of ncRNA biology, we have developed a proprietary, modular pipeline for a client designed to identify and prioritize high-confidence ncRNA targets across diverse disease contexts. This system enables consistent and scalable analysis while integrating multiple layers of biological evidence. Built with flexibility and reproducibility in mind, the pipeline reflects our deep understanding of both the molecular nuances and practical challenges involved in ncRNA research.
Impact of RNA-Centered Innovation
More than a data analysis tool, this framework supports informed decision-making by translating complex molecular data into actionable insights. The therapeutic relevance of ncRNAs is already evident in several established examples, underscoring the importance and translational potential of ncRNA research.
While siRNA drugs targeting mRNAs are FDA-approved, therapeutics that directly target endogenous ncRNAs are still under research. However, several candidates have already reached clinical development, highlighting the therapeutic potential of this class of molecules. Some notable cases include:
miR-122: Miravirsen
An antisense oligonucleotide designed to inhibit liver-specific miR-122, a key host factor in hepatitis C virus replication. Miravirsen reached Phase II clinical trials, showing dose-dependent reductions in viral load and demonstrating the feasibility of miRNA-targeted therapies. Reference: In Vitro Antiviral Activity and Preclinical and Clinical Resistance Profile of Miravirsen, a Novel Anti-Hepatitis C Virus Therapeutic Targeting the Human Factor miR-122 | Antimicrobial Agents and Chemotherapy
lncRNA MALAT
Preclinical studies have shown that silencing MALAT1 via antisense oligonucleotides can reduce metastasis in various cancers, including triple-negative breast cancer. MALAT1 remains one of the most studied and promising lncRNA targets for oncology. Reference: Abstract 1541: LncRNA MALAT1 as a potential therapeutic target in triple negative breast cancer | Cancer Research | American Association for Cancer Research
lncRNA H19: DTA-H19
A plasmid-based therapy designed to target the H19 promoter for tumor-specific expression of a diphtheria toxin. It has entered early clinical trials for bladder and ovarian cancer. Reference: Frontiers | The role of lncRNA H19 in tumorigenesis and drug resistance of human Cancers
circRNAs as miRNA sponges
Synthetic circular RNAs have been engineered to sponge oncogenic miRNAs such as miR-21 and miR-93 in cancer models, showing promising preclinical results in modulating tumor growth and gene expression. Reference: Artificial Circular RNA Sponges Targeting MicroRNAs as a Novel Tool in Molecular Biology: Molecular Therapy Nucleic Acids
These examples illustrate that ncRNAs can serve as effective therapeutic targets or tools. However, an efficient identification such candidates remains a complex task. Our pipeline addresses this challenge by integrating diverse data types to systematically prioritize ncRNAs with high therapeutic potential. By streamlining this process, the framework helps accelerate the transition from data to discovery, ultimately supporting the development of future RNA-based therapeutics.